

coli DNA polymerase I by the action of the enzyme protease subtilisin. The Klenow fragment is the result of the proteolytic digestion of E. DNA polymerase V takes part in DNA repair. On the contrary, DNA polymerase IV takes part in non-targeted mutagenesis. Meanwhile, DNA polymerase III holoenzyme is the main enzyme responsible for DNA replication in prokaryotes. Additionally, DNA polymerase II only has 3′ to 5′ exonuclease activity. Here, DNA polymerase I has both 3′ to 5′ and 5′ to 3′ exonuclease activity. Moreover, the main types of DNA polymerases that occur in prokaryotes include DNA polymerase I, II, III, IV, and V. Other DNA polymerases take part in DNA repair. However, only α, δ, and ε polymerases take part in DNA replication. Generally, in eukaryotes, six types of DNA polymerases occur they are DNA polymerases α, β, γ, δ, ε, and ζ. Some of the significant features of DNA polymerase include the addition of nucleotides in the 5′ to 3′ direction on the growing strand and the requirement of an RNA primer – in other words, a pre-existing 3′-OH group for the initiation of synthesis.
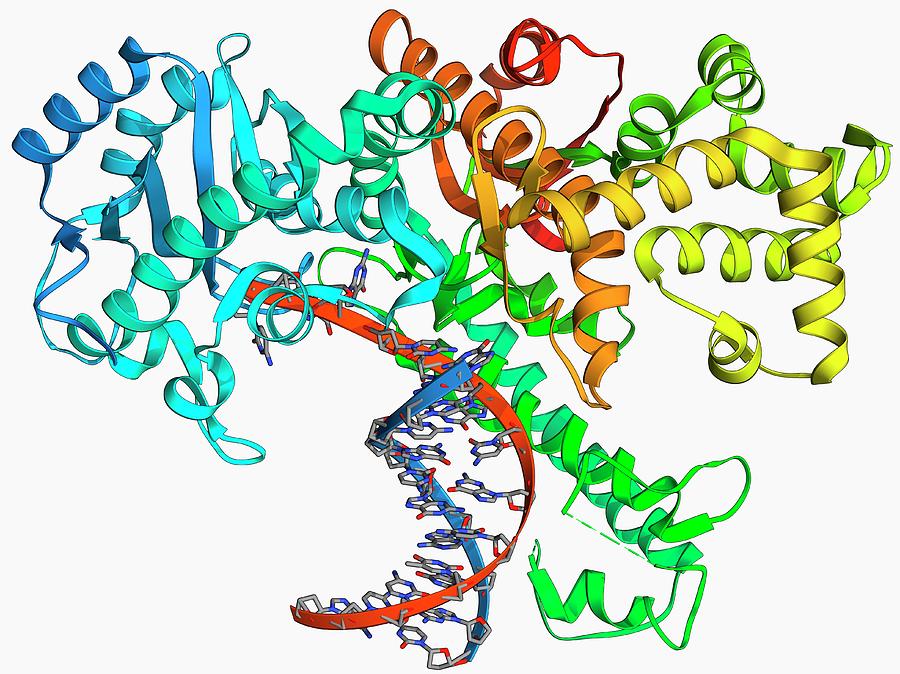
– Comparison of Key Differences Key TermsĭNA Polymerase, Klenow Fragment What is DNA PolymeraseĭNA polymerase is the enzyme responsible for the synthesis of DNA inside the cell. Difference Between DNA Polymerase and Klenow Fragment Similarities Between DNA Polymerase and Klenow FragmentĤ. Ollis, D.L., Brick, P., Hamlin, R., Xuong, N.G., Steitz, T.A.ĭepartment of Molecular Biophysics, Yale University, New Haven, Connecticut 06511.The main difference between DNA polymerase and Klenow fragment is that DNA polymerase is the enzyme that catalyzes the synthesis of DNA molecules, whereas the Klenow fragment is the largest segment of DNA polymerase I obtained by the enzymatic cleavage of protease subtilisin.ĭNA polymerase and Klenow fragment are two types of protein components that involve in the synthesis of DNA molecules. Structure of Large Fragment of Escherichia Coli DNA Polymerase I Complexed with dTMP.Co-Crystal Structure of an Editing Complex of Klenow Fragment with DNAįreemont, P.S., Friedman, J.M., Beese, L.S., Steitz, T.A.Coli DNA Polymerase F: A Two Metal Ion Mechanism

Structural Basis for the 3'-5' Exonuclease Activity of E.Structure of DNA Polymerase I Klenow Fragment Bound to Duplex DNAīeese, L.S., Derbyshire, V., Steitz, T.A.However, we conclude that the position of at least the dNMP moiety of dNTP in the binary complex is not likely to be the same as in its catalytically relevant complex with primer-template DNA. The dNTP binding site observed in the crystal is consistent with the results of chemical modification including cross-linking and is also near many of the amino acid residues whose mutation affects catalysis.
KLENOW FRAGMENT MOLECULAR WEIGHT PLUS
The dNTP binds adjacent to the O-helix with its triphosphate moiety anchored by three positively charged residues, Arg 754, Arg 682, and Lys 758, plus His 734 and Gln 708. Diversity, Equity, Inclusion, and AccessĬrystal structures of the Klenow fragment (KF) of DNA polymerase I from Escherichia coli complexed with deoxynucleoside triphosphate (dNTP) or with pyrophosphate (PPi) determined to 3.9-A resolution by X-ray crystallography show these molecules binding within the cleft of the polymerase domain and surrounded by residues previously implicated in dNTP binding.Citation, Usage, Privacy Policies, Logo.Biologically Interesting Molecule Reference Dictionary (BIRD).


 0 kommentar(er)
0 kommentar(er)
